The Sonic Hedgehog (Shh) signalling pathway performs a number of roles throughout embryonic growth and underneath pathological situations. Although the core elements of the Shh pathway are conserved, the regulation of sign transduction varies considerably amongst species and cell sorts.
Protein kinases Ulk3 and Pka are concerned in the Shh pathway as modulators of the actions of Gli transcription elements, that are the nuclear mediators of the sign. Here, we examine the regulation and actions of two GLI1 isoforms, full-length GLI1 (GLI1FL) and GLI1ΔN. The latter protein lacks the first 128 amino acids together with the conserved phosphorylation cluster and the binding motif for SUFU, the key regulator of GLI exercise. Both GLI1 isoforms are co-expressed in all human cell traces analysed and possess related DNA binding exercise.
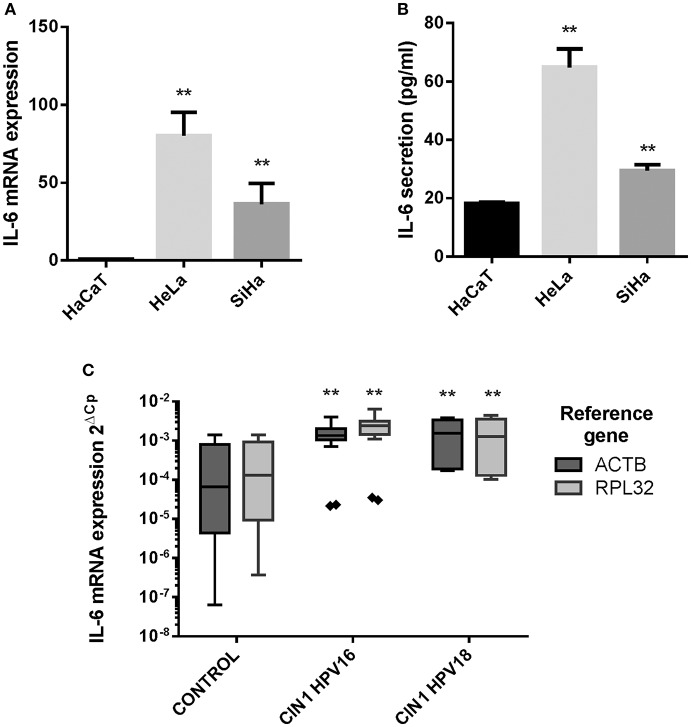
ULK3 potentiates the transcriptional exercise of each GLI1 proteins, whereas PKA inhibits the exercise of GLI1ΔN, however not GLI1FL. In addition to its well-established function as a transcriptional activator, GLI1FL acts as a repressor by inhibiting transcription from the early promoters of human papillomavirus kind 18 (HPV18).
Additionally, in comparison with GLI1ΔN, GLI1FL is a stronger suppressor of replication of a number of HPV sorts. Altogether, our knowledge present that the N-terminal half of GLI1FL is essential for the realization of its full potential as a transcriptional regulator.
A CRISPR Test for Detection of Circulating Nuclei Acids
Emerging CRISPR-based nucleic acid detection reveals nice promise in molecular prognosis of illnesses. CRISPR-Cas12a can sensitively and particularly detect human papillomavirus (HPV) DNA in anal swabs.
However, the present CRISPR-Cas12a system wants auxiliary and costly tools, which restrict its utility as a point-of-care (POC) diagnostic instrument. This research aimed to develop CRISPR-Cas12a as a POC take a look at to straight goal plasma for circulating HPV DNA detection by instantly studying outcomes with bare eyes.
Cell-cultured supernatants of both HPV16- or 18-positive most cancers cells have been handled with lysis buffer adopted by isothermal amplification with out DNA isolation. Cas12a, crRNA, and fluorescent-biotin reporters have been incubated with the lysates.
Our knowledge confirmed that integrating CRISPR-Cas12a with lateral-flow strips might straight and particularly detect HPV16 and 18 in the liquid samples with the similar restrict of detection (0.24 fM) as did polymerase chain response however requiring much less time.
Furthermore, the CRISPR-Cas12a system might quickly detect presence of HPV16 and HPV18 in plasma samples of 13 of 14 and 3 of 10 the sufferers with histopathological prognosis of cervical most cancers, respectively.
Therefore, a CRISPR-Cas12a-based POC system was developed for conveniently detecting circulating nuclei acid targets in physique fluids with out requiring technical experience and ancillary machineries.
Humanpapillomavirus (HPV) 16/18 genotyping is an efficient technique for triage of high-risk (hr) HPV-positive girls in major hrHPV screening for cervical most cancers. The current research aimed to judge whether or not co-infected with different hrHPV sorts will have an effect on the threat of cervical carcinogenesis in HPV16/18 constructive girls.
A complete of 313,704 girls aged ≥30 years have been screened in China. Among them, 4,933 HPV16/18-positive individuals underwent colposcopy-directed biopsy. The HPV genotypes have been recognized utilizing the Cobas HPV genotyping system. Multinomial logistic regression was used to mannequin completely different HPV16/18 an infection patterns.
The general prevalence charges of hrHPV and HPV16/18 have been 7.85% (24,456/311,382) and 1.95% (6,086/311,382) respectively. Among HPV16/18 constructive people, 33.24% (2,023/6,086) have been co-infection with a number of sorts. Of the 4933 girls who underwent colposcopy, their HPV16/18 an infection patterns have been as follows: 52.38% (2,584/4,933) HVP16 solely, 23.54% (1,161/4,933) HPV16 + different hrHPVs, 14.98% (739/4,933) HPV18 solely, 6.83% (337/4,933) HPV18 + different hrHPVs, 1.13% (56/4,933) HPV16 + 18, 1.13% (56/4,933) HPV16 + 18+different hrHPVs.
After adjusting for cofactors, in contrast with single HPV16 an infection, the threat of creating cervical intraepithelial neoplasia (CIN) grade Three or better (CIN3+) was considerably decrease in HPV16 + different hrHPVs group (odds ratio [OR] = 0.637, 95% confidence interval [CI] = 0.493-0.822).HPV16/18 co-infection with different hrHPVs is a typical phenomenon.
Different HPV16/18 an infection patterns might affect the threat of cervical carcinogenesis. HPV16 co-infected with different hrHPVs seems to have a decrease related threat of CIN3+ in ≥30 years outdated girls.
Analytical Evaluation of the Human Papillomavirus HPV DNA Array E1-Based Genotyping Assay
Cervical most cancers is attributable to a persistent an infection of humanpapillomavirus (HPV). Therefore, exams which detect the carcinogenic virus can be utilized for cervical most cancers screening.
This is the first analysis of the HPV DNA Array (AID Diagnostika, Strassberg, Germany), an E1-based genotyping polymerase chain response (PCR) take a look at for identification of 29 HPV sorts (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 67, 68, 69, 70, 73, 82, 85, and 97).Analytical efficiency of the assay was assessed with cervical most cancers cell traces with recognized HPV standing, and preselected scientific cervical scrapings genotyped by multiplexed genotyping (MPG) with a Luminex readout (validated in-house assay).
Intra- and inter-laboratory reproducibility experiments have been carried out to make sure the reliability of the assay.HPV DNA Array recognized the intrinsic HPV genotype in all cervical most cancers cell traces and demonstrated a excessive sensitivity for HPV16 probe (1 cell per PCR response), in addition to HPV18 and 45 probes (100 cells per PCR response).
When in contrast with MPG, HPV DNA Array confirmed a great settlement of 92.2% for HPV detection irrespective of kind (κ = 0.601), and demonstrated excessive settlement for HPV16 (80.7%, κ = 0.836) and HPV18 (86.7%, κ = 0.925). Furthermore, excessive intra-/inter-laboratory reproducibility was noticed (90.9-100%).HPV DNA Array confirmed excessive sensitivity for proper HPV genotype detection in experimental and scientific samples with a great correlation to the reference take a look at.
Since HPV DNA Array is predicated on a easy multiplexed PCR adopted by reverse hybridization in a 96-well format and automated visible readout by AID ELISpot reader, it’s succesful of excessive throughput in a time-effective method. HPV DNA Array could possibly be thought-about for prolonged HPV genotyping of cervical smears.
