The E6 protein from high-risk human papillomavirus varieties interacts with and degrades a number of PDZ domain-containing proteins that localize to adherens junctions or tight junctions in polarized epithelial cells. We have recognized the tight junction-associated multi-PDZ protein PATJ (PALS1-associated TJ protein) as a novel binding associate and degradation target of high-risk varieties 16 and 18 E6.
PATJ capabilities in the meeting of the evolutionarily conserved CRB-PALS1-PATJ and Par6-aPKC-Par3 complexes and is crucial for the formation of tight junctions in polarized cells.
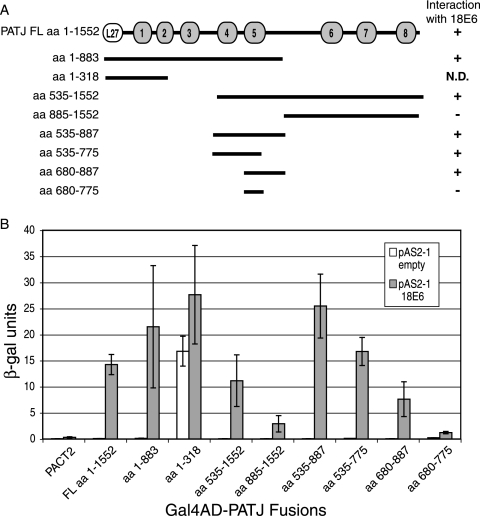
The means of sort 18 E6 full-length to bind to, and the subsequent degradation of, PATJ is depending on its C-terminal PDZ binding motif. We display that the spliced 18 E6* protein, which lacks a C-terminal PDZ binding motif, associates with and degrades PATJ independently of full-length 18 E6.
Thus, PATJ is the first binding associate that is degraded in response to each isoforms of 18 E6. The means of E6 to make the most of a non-E6AP ubiquitin ligase for the degradation of a number of PDZ binding companions has been recommended. We additionally display that 18 E6-mediated degradation of PATJ is not inhibited in cells the place E6AP is silenced by shRNA. This means that the E6-E6AP complicated is not required for the degradation of this protein target.
Evolutionary dynamics of variant genomes of human papillomavirus varieties 18, 45, and 97.
Human papillomavirus sort 18 (HPV18) and HPV45 account for about 20% of all cervix cancers. We present that HPV18, HPV45, and the just lately found HPV97 comprise a clade sharing a most up-to-date widespread ancestor inside HPV alpha7 species.
Variant lineages of these HPV varieties had been labeled by sequence evaluation of the upstream regulatory area/E6 area amongst cervical samples from a population-based research in Costa Rica, and 27 consultant genomes from every main variant lineage had been sequenced.
Nucleotide variation inside HPV18 and HPV45 was 3.82% and 2.39%, respectively, and amino acid variation was 4.73% and 2.87%, respectively. Only 18 nucleotide variations, of which 10 had been nonsynonymous, had been recognized amongst three HPV97 genomes. Full-genome comparisons revealed maximal variety between HPV18 African and non-African variants (2.6% dissimilarity), whereas HPV18 Asian-American [E1 (AA)] and European (E2) variants had been intently associated (lower than 0.5% dissimilarity); HPV45 genomes had a maximal distinction of 1.6% nucleotides.
Using a Bayesian Markov chain Monte Carlo (MCMC) methodology, the divergence instances of HPV18, -45, and -97 from their most up-to-date widespread ancestors indicated that HPV18 diverged roughly 7.7 million years (Myr) in the past, whereas HPV45 and HPV97 cut up off round 5.7 Myr in the past, in a interval encompassing the divergence of the nice ape species. Variants inside the HPV18/45/97 lineages had been estimated to have diverged from their widespread ancestors in the genus Homo inside the final 1 Myr (<0.7 Myr).
To examine the molecular foundation of HPV18, HPV45, and HPV97 evolution, regression fashions of codon substitution had been used to determine lineages and amino acid websites beneath selective strain. The E5 open studying body (ORF) of HPV18 and the E4 ORFs of HPV18, HPV45, and HPV18/45/97 had nonsynonymous/synonymous substitution charge ratios (d(N)/d(S)) over 1 indicative of optimistic Darwinian choice.
The L1 ORF of HPV18 genomes had an elevated proportion of nonsynonymous substitutions (4.93%; common d(N)/d(S) ratio [M3] = 0.3356) in comparison with HPV45 (1.86%; M3 = 0.1268) and HPV16 (2.26%; M3 = 0.1330) L1 ORFs.
In distinction, HPV18 and HPV16 genomes had comparable amino acid substitution charges inside the E1 ORF (2.89% and 3.24%, respectively), whereas HPV45 E1 was extremely conserved (amino acid substitution charge was 0.77%).
These information present an evolutionary historical past of this medically vital clade of HPVs and determine an surprising divergence of the L1 gene of HPV18 which will have scientific implications for the long-term use of an L1-virus-like particle-based prophylactic vaccine.
